Developing a 3D deep learning pipeline for segmenting neuronal somas in whole-brain light sheet fluorescence microscopy rodent data.
Supervised by Prof. Maged Goubran, funded by 2023 T-CAIREM AI in Medicine Summer Research Studentship Program.
Mapping neuronal cell bodies is important to study how the brain functions, understand the machanims underlying neurodiseases, and also develop targeted interventions for neurodegenerative diseases. Mouse, often used as an important model organism, have 8–14 million of neurons that are in the scale of micrometres, how can people study them?
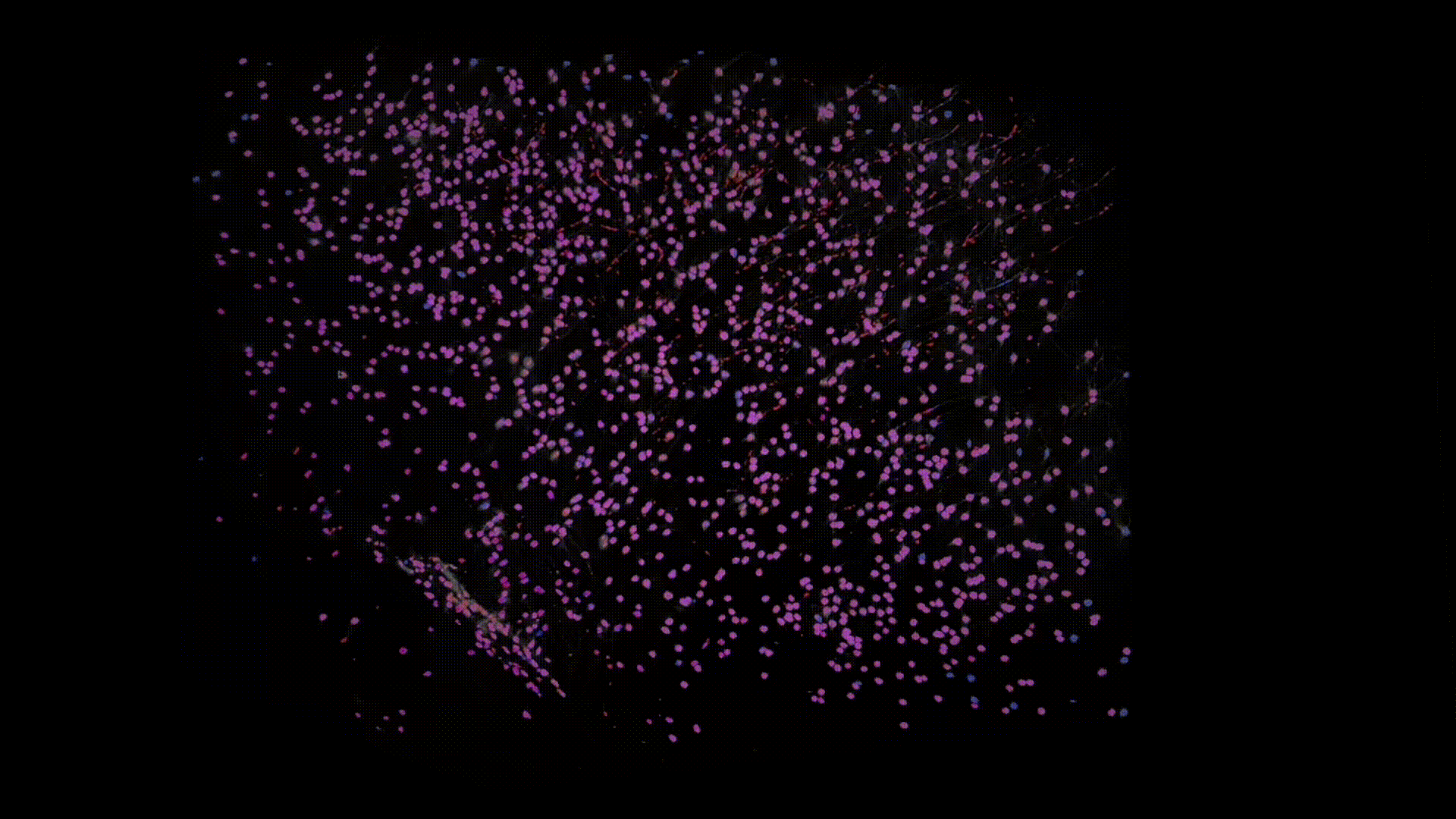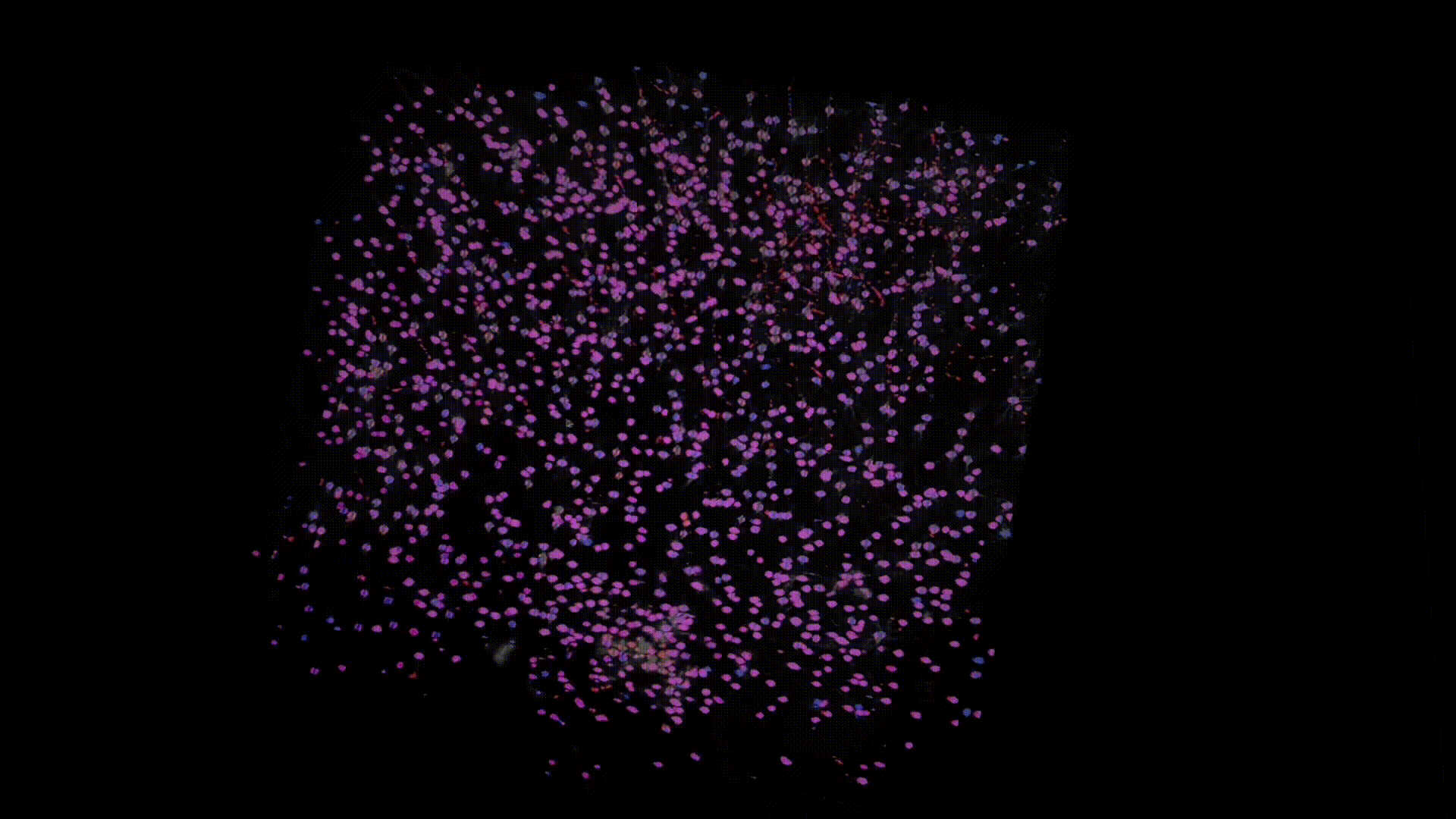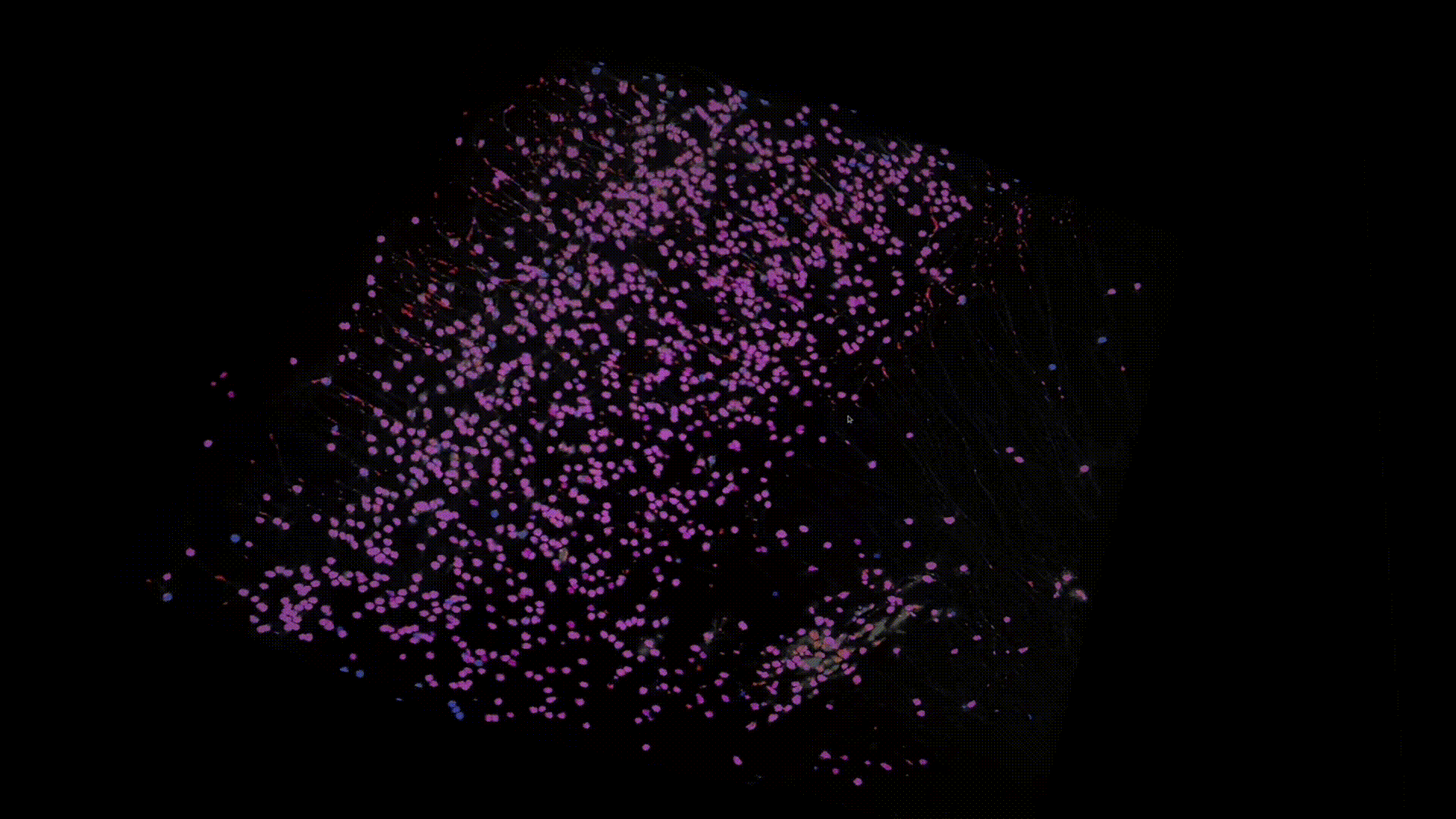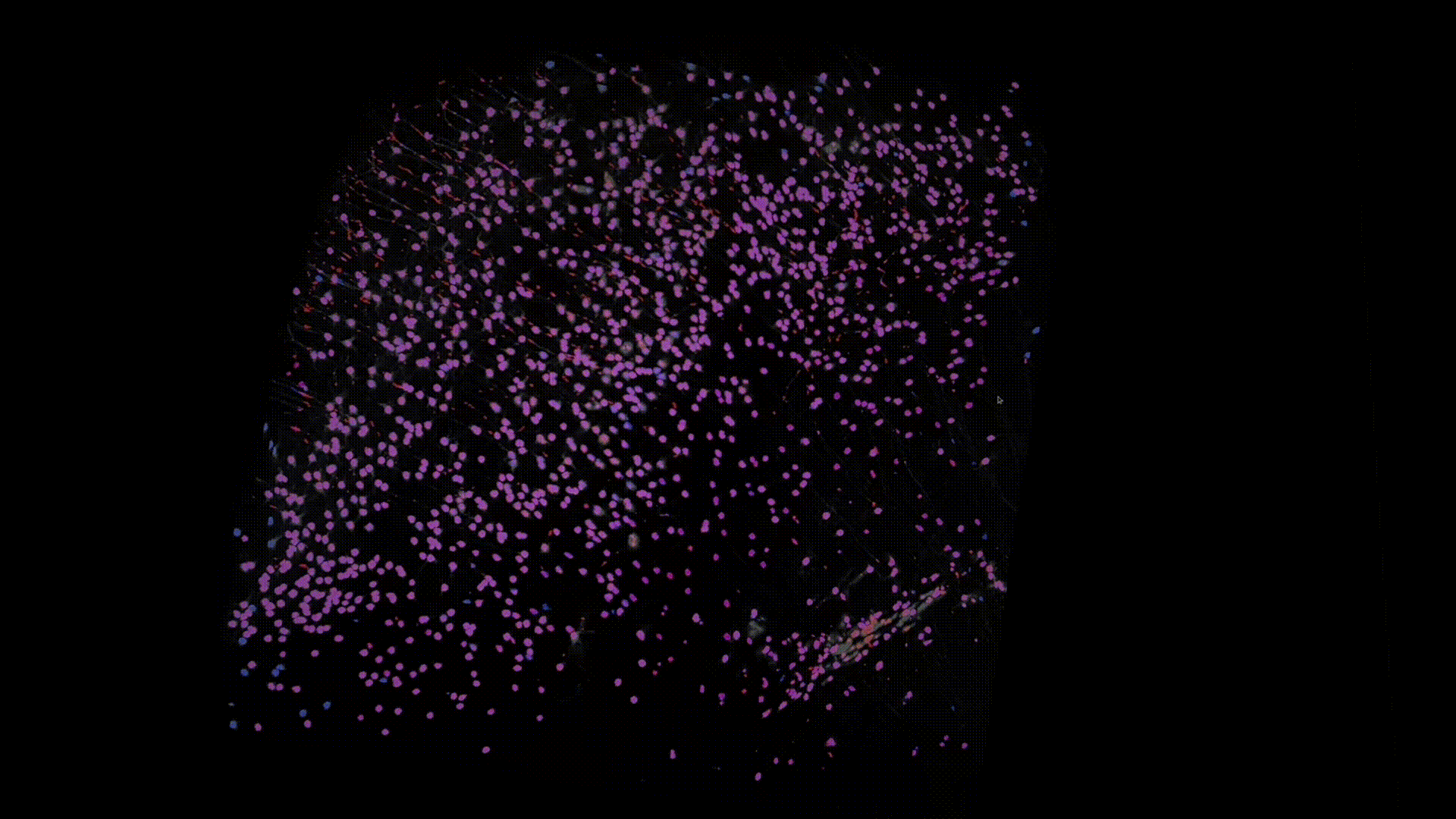
Thanks to recent advances in tissue clearing technology and light sheet fluorescence microscopy (LSFM) imaging techniques, we can obtain a transparent whole brain image of neuronal cell bodies in mice. Below is an example of the data that I have worked with.
As current computational pipelines for identifying the neurons are either 2D-based, or have problems such as low speed, lack of generalizability, and requirement of lots of human intervention. In this project, I have been working on developing an automatic 3D deep learning-based computational pipeline to segment neuron cell bodies in whole-brain LSFM image data.
The model I am working on is based on U-Net with some modifications that we specifically designed for dealing with 3D LSTM image data. Here are some preliminary results. Below I overlay the model prediction (in blue) and the ground-truth (in red). Note that the branches, colored in red are actually axons not neuron somas, which are mislabeled in groundtruth, whereas our model successfully avoids them.