publications
publications in reversed chronological order.
2025
- Ahmadreza Attarpour, Jonas Osmann, Anthony Rinaldi, and 14 more authorsNature Methods, Jan 2025
Teravoxel-scale, cellular-resolution images of cleared rodent brains acquired with light-sheet fluorescence microscopy have transformed the way we study the brain. Realizing the potential of this technology requires computational pipelines that generalize across experimental protocols and map neuronal activity at the laminar and subpopulation-specific levels, beyond atlas-defined regions. Here, we present artficial intelligence-based cartography of ensembles (ACE), an end-to-end pipeline that employs three-dimensional deep learning segmentation models and advanced cluster-wise statistical algorithms, to enable unbiased mapping of local neuronal activity and connectivity. Validation against state-of-the-art segmentation and detection methods on unseen datasets demonstrated ACE’s high generalizability and performance. Applying ACE in two distinct neurobiological contexts, we discovered subregional effects missed by existing atlas-based analyses and showcase ACE’s ability to reveal localized or laminar neuronal activity brain-wide. Our open-source pipeline enables whole-brain mapping of neuronal ensembles at a high level of precision across a wide range of neuroscientific applications.
2024
- Fengqing Yu, Ting S. Li, Joshua S. Speagle, and 9 more authorsThe Astrophysical Journal, Oct 2024
Blue horizontal branch stars (BHBs), excellent distant tracers for probing the Milky Way’s halo density profile, are distinguished in the versus (i − z)0 color space from another class of stars, blue straggler stars. We develop a Bayesian mixture model to classify BHBs using high-precision photometry data from the Dark Energy Survey Data Release 2 (DES DR2). We select ∼2100 highly probable BHBs based on their griz photometry and the associated uncertainties, and we use these stars to map the stellar halo over the Galactocentric radial range 20 kpc ≲ R ≲ 70 kpc. After excluding known stellar overdensities, we find that the number density n ⋆ of BHBs can be represented by a power-law density profile n ⋆ ∝ R −α with an index of , consistent with existing literature values. In addition, we examine the impact of systematic errors and the spatial inhomogeneity on the fitted density profile. Our work demonstrates the effectiveness of high-precision griz photometry in selecting BHBs. The upcoming photometric survey from the Rubin Observatory, expected to reach depths 2–3 mag greater than DES during its 10 yr mission, will enable us to investigate the density profile of the Milky Way’s halo out to the virial radius, unraveling the complex processes of formation and evolution in our Galaxy.
2023
-
 Ethan Hau Yin Lam, Fengqing Yu, Sabrina Zhu, and 1 more authorInternational Journal of Molecular Sciences, Oct 2023
Ethan Hau Yin Lam, Fengqing Yu, Sabrina Zhu, and 1 more authorInternational Journal of Molecular Sciences, Oct 2023In the past decade, immense progress has been made in advancing personalized medicine to effectively address patient-specific disease complexities in order to develop individualized treatment strategies. In particular, the emergence of 3D bioprinting for in vitro models of tissue and organ engineering presents novel opportunities to improve personalized medicine. However, the existing bioprinted constructs are not yet able to fulfill the ultimate goal: an anatomically realistic organ with mature biological functions. Current bioprinting approaches have technical challenges in terms of precise cell deposition, effective differentiation, proper vascularization, and innervation. This review introduces the principles and realizations of bioprinting with a strong focus on the predominant techniques, including extrusion printing and digital light processing (DLP). We further discussed the applications of bioprinted constructs, including the engraftment of stem cells as personalized implants for regenerative medicine and in vitro high-throughput drug development models for drug discovery. While no one-size-fits-all approach to bioprinting has emerged, the rapid progress and promising results of preliminary studies have demonstrated that bioprinting could serve as an empowering technology to resolve critical challenges in personalized medicine.
-
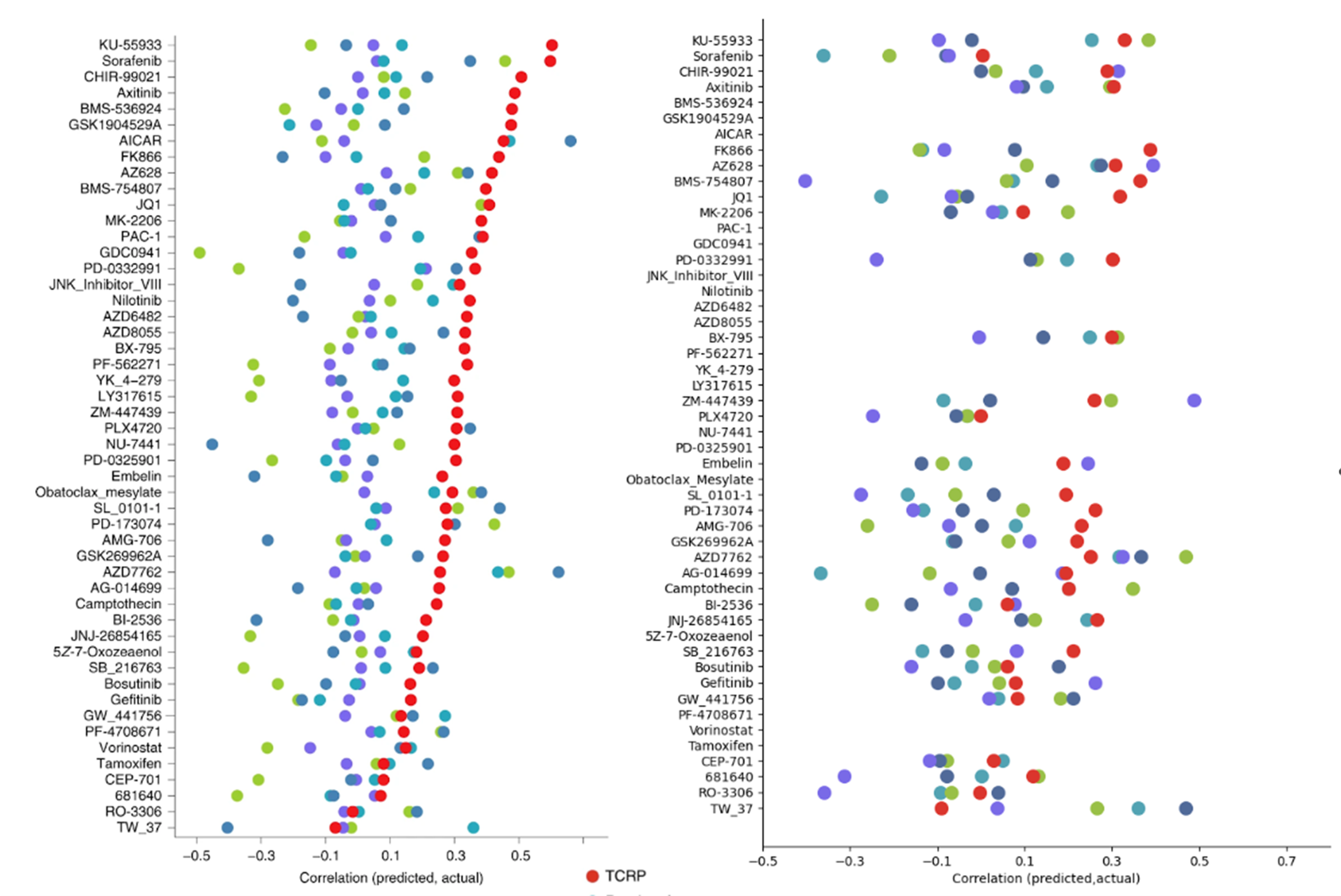 Emily So, Fengqing Yu, Bo Wang, and 1 more authorNature Machine Intelligence, Jul 2023
Emily So, Fengqing Yu, Bo Wang, and 1 more authorNature Machine Intelligence, Jul 2023Machine learning and artificial intelligence methods are increasingly being used in personalized medicine, including precision oncology. Ma et al. (Nature Cancer 2021) have developed a new method called ‘transfer of cell line response prediction’ (TCRP) to train predictors of drug response in cancer cell lines and optimize their performance in higher complex cancer model systems via few-shot learning. TCRP has been presented as a successful modelling approach in multiple case studies. Given the importance of this approach for assisting clinicians in their treatment decision processes, we sought to independently reproduce the authors’ findings and improve the reusability of TCRP in new case studies, including validation in clinical-trial datasets—a high bar for drug-response prediction. Our reproducibility results, while not reaching the same level of superiority as those of the original authors, were able to confirm the superiority of TCRP in the original clinical context. Our reusability results indicate that, in the majority of novel clinical contexts, TCRP remains the superior method for predicting response for both preclinical and clinical settings. Our results thus support the superiority of TCRP over established statistical and machine learning approaches in preclinical and clinical settings. We also developed new resources to increase the reusability of the TCRP model for future improvements and validation studies.